Regulation of Abscisic Acid Biosynthesis1

Update on Abscisic Acid Biosynthesis Regulation
Regulation of Abscisic Acid Biosynthesis1
Liming Xiong*and Jian-Kang Zhu
Donald Danforth Plant Science Center,975North Warson Road,St.Louis,Missouri63132(L.X.);and Department of Plant Sciences,University of Arizona,Tucson,Arizona85721(J.-K.Z.)
Plant growth and development are regulated by internal signals and by external environmental con-ditions.One important regulator that coordinates growth and development with responses to the en-vironment is the sesquiterpenoid hormone abscisic acid(ABA).ABA plays important roles in many cel-lular processes including seed development,dor-mancy,germination,vegetative growth,and environ-mental stress responses.These diverse functions of ABA involve complex regulatory mechanisms that control its production,degradation,signal percep-tion,and transduction.Because of the key role of ABA in plant stress responses,understanding these regulatory mechanisms will help devise rational strategies to breed or genetically engineer crop plants with increased tolerance to adverse environmental conditions.
Since the discovery of ABA in the early1960s, much effort has been devoted to understanding how ABA is synthesized.Through genetic and biochemi-cal studies,the pathway for ABA biosynthesis in higher plants is now understood in great detail.Re-cently,all the major genes for the enzymes in the biosynthesis pathway have been identified(Schwartz et al.,2003).The new challenge is to understand how these biosynthesis genes,and the biosynthetic path-way as a whole,are regulated.Although much re-mains to be learned about the regulatory mechanism, evidence thus far indicates that ABA biosynthesis is subject to complex regulation during plant develop-ment and in response to environmental stresses.In this Update,we first present a brief overview of the functions of ABA and the biosynthesis pathway.We then focus on the regulation of ABA production and attempt to provide some future directions in ABA biosynthesis studies.
BIOLOGICAL FUNCTIONS OF ABA
Under non-stressful conditions,ABA in plant cells is maintained at low levels.Some low levels of ABA may be required for normal plant growth,as evi-denced by reduced vigor observed in ABA-deficient mutant plants that can be restored to the wild-type level of growth by exogenous ABA(Finkelstein and Rock,2002).Because all ABA-deficient mutants still have certain basal levels of ABA that are not dramat-ically lower than those in the wild type under normal growth conditions,it is difficult to uncover the cel-lular processes that require a very small amount of ABA.As a consequence,our knowledge of ABA functions has been gained mainly from observations with ABA at elevated levels,either from endogenous or exogenous sources.ABA levels can increase dra-matically during seed maturation and in response to environmental stresses.Thus,ABA functions have been most extensively studied in these two processes. During seed development,ABA is known to initi-ate the following programs:embryo maturation,syn-thesis of storage reserves and late embryogenesis-abundant(LEA)proteins,and initiation of seed dormancy,although ABA is not the sole regulator of these processes.In particular,the induction of LEA protein synthesis to preserve the viability of embryos in the extremely dry condition of seeds is related to the role of ABA in promoting synthesis of LEA-like proteins in vegetative tissues to tolerate dehydration stress.Embryos from ABA antibody-expressing plants lose their viability as a result of desiccation intolerance(Phillips et al.,1997).
In vegetative tissues,ABA levels increase when plants encounter adverse environmental conditions such as drought,salt,and to a lesser extent,low temperatures.Although a higher level of exogenous ABA inhibits plant growth under non-stressful con-ditions,an increased ABA content is beneficial for plants under environmental stress as a result of ABA-induced changes at the cellular and whole-plant lev-els.ABA promotes the closure of stomata to mini-mize transpirational water loss.It also mitigates stress damage through the activation of many stress-responsive genes that encode enzymes for the bio-synthesis of compatible osmolytes and LEA-like pro-teins,which collectively increase plant stress tolerance(Hasegawa et al.,2000;Bray,2002;Finkel-stein et al.,2002).In addition,ABA has been shown to offset the inhibitory effect of stress-induced ethyl-ene on plant growth(Sharp,2002).Plant mutants defective in ABA biosynthesis are more susceptible to the environmental stresses and have been isolated in stress sensitivity screens(Xiong et al.,2002b).Im-portantly,manipulating ABA levels by changing the
1This work was supported by grants from the National Science Foundation,the National Institutes of Health,and the U.S.Depart-ment of Agriculture.
*Corresponding author;e-mail lxiong@https://www.wendangku.net/doc/bb4899823.html,;fax 314–587–1562.
https://www.wendangku.net/doc/bb4899823.html,/cgi/doi/10.1104/pp.103.025395.
expression of key ABA biosynthetic genes provides an effective means to increase plant stress resistance.
THE ABA BIOSYNTHESIS PATHWAY
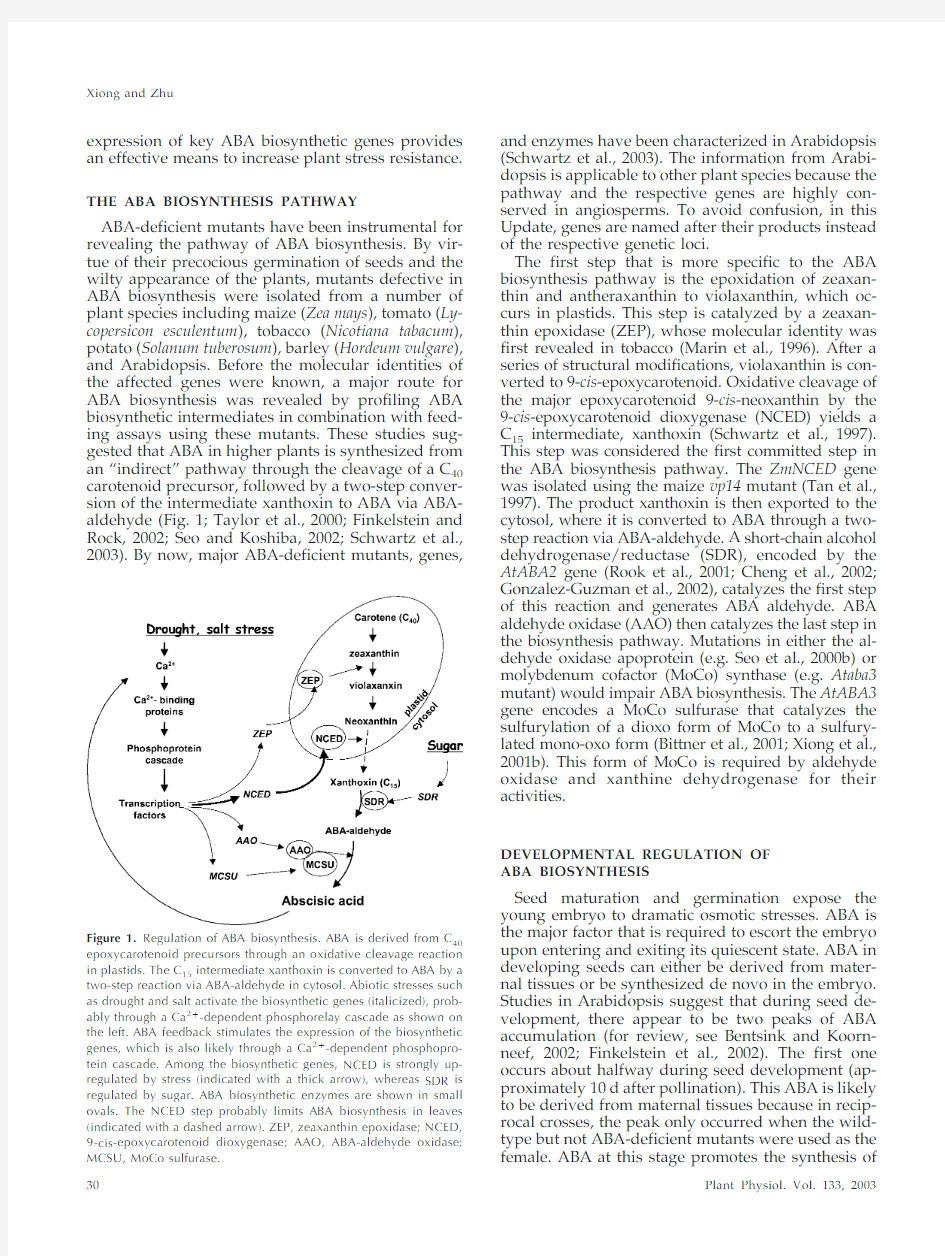
ABA-deficient mutants have been instrumental for revealing the pathway of ABA biosynthesis.By vir-tue of their precocious germination of seeds and the wilty appearance of the plants,mutants defective in ABA biosynthesis were isolated from a number of plant species including maize (Zea mays ),tomato (Ly-copersicon esculentum ),tobacco (Nicotiana tabacum ),potato (Solanum tuberosum ),barley (Hordeum vulgare ),and Arabidopsis.Before the molecular identities of the affected genes were known,a major route for ABA biosynthesis was revealed by profiling ABA biosynthetic intermediates in combination with feed-ing assays using these mutants.These studies sug-gested that ABA in higher plants is synthesized from an “indirect ”pathway through the cleavage of a C 40carotenoid precursor,followed by a two-step conver-sion of the intermediate xanthoxin to ABA via ABA-aldehyde (Fig.1;Taylor et al.,2000;Finkelstein and Rock,2002;Seo and Koshiba,2002;Schwartz et al.,2003).By now,major ABA-deficient mutants,genes,
and enzymes have been characterized in Arabidopsis (Schwartz et al.,2003).The information from Arabi-dopsis is applicable to other plant species because the pathway and the respective genes are highly con-served in angiosperms.To avoid confusion,in this Update,genes are named after their products instead of the respective genetic loci.
The first step that is more specific to the ABA biosynthesis pathway is the epoxidation of zeaxan-thin and antheraxanthin to violaxanthin,which oc-curs in plastids.This step is catalyzed by a zeaxan-thin epoxidase (ZEP),whose molecular identity was first revealed in tobacco (Marin et al.,1996).After a series of structural modifications,violaxanthin is con-verted to 9-cis -epoxycarotenoid.Oxidative cleavage of the major epoxycarotenoid 9-cis -neoxanthin by the 9-cis -epoxycarotenoid dioxygenase (NCED)yields a C 15intermediate,xanthoxin (Schwartz et al.,1997).This step was considered the first committed step in the ABA biosynthesis pathway.The ZmNCED gene was isolated using the maize vp14mutant (Tan et al.,1997).The product xanthoxin is then exported to the cytosol,where it is converted to ABA through a two-step reaction via ABA-aldehyde.A short-chain alcohol dehydrogenase/reductase (SDR),encoded by the AtABA2gene (Rook et al.,2001;Cheng et al.,2002;Gonzalez-Guzman et al.,2002),catalyzes the first step of this reaction and generates ABA aldehyde.ABA aldehyde oxidase (AAO)then catalyzes the last step in the biosynthesis pathway.Mutations in either the al-dehyde oxidase apoprotein (e.g.Seo et al.,2000b)or molybdenum cofactor (MoCo)synthase (e.g.Ataba3mutant)would impair ABA biosynthesis.The AtABA3gene encodes a MoCo sulfurase that catalyzes the sulfurylation of a dioxo form of MoCo to a sulfury-lated mono-oxo form (Bittner et al.,2001;Xiong et al.,2001b).This form of MoCo is required by aldehyde oxidase and xanthine dehydrogenase for their activities.
DEVELOPMENTAL REGULATION OF ABA BIOSYNTHESIS
Seed maturation and germination expose the young embryo to dramatic osmotic stresses.ABA is the major factor that is required to escort the embryo upon entering and exiting its quiescent state.ABA in developing seeds can either be derived from mater-nal tissues or be synthesized de novo in the embryo.Studies in Arabidopsis suggest that during seed de-velopment,there appear to be two peaks of ABA accumulation (for review,see Bentsink and Koorn-neef,2002;Finkelstein et al.,2002).The first one occurs about halfway during seed development (ap-proximately 10d after pollination).This ABA is likely to be derived from maternal tissues because in recip-rocal crosses,the peak only occurred when the wild-type but not ABA-deficient mutants were used as the female.ABA at this stage promotes the synthesis
of
Figure 1.Regulation of ABA biosynthesis.ABA is derived from C 40epoxycarotenoid precursors through an oxidative cleavage reaction in plastids.The C 15intermediate xanthoxin is converted to ABA by a two-step reaction via ABA-aldehyde in cytosol.Abiotic stresses such as drought and salt activate the biosynthetic genes (italicized),prob-ably through a Ca 2?-dependent phosphorelay cascade as shown on the left.ABA feedback stimulates the expression of the biosynthetic genes,which is also likely through a Ca 2?-dependent phosphopro-tein cascade.Among the biosynthetic genes,NCED is strongly up-regulated by stress (indicated with a thick arrow),whereas SDR is regulated by sugar.ABA biosynthetic enzymes are shown in small ovals.The NCED step probably limits ABA biosynthesis in leaves (indicated with a dashed arrow).ZEP,zeaxanthin epoxidase;NCED,9-cis -epoxycarotenoid dioxygenase;AAO,ABA-aldehyde oxidase;MCSU,MoCo sulfurase.Xiong and Zhu
storage proteins.Embryos from ABA antibody-expressing plants either did not accumulate or accu-mulated a much lower level of storage proteins rel-ative to the wild-type embryos(Phillips et al.,1997). The second peak with less significant ABA accumu-lation(about one-third of the first peak)is from bio-synthesis in the embryo and may activate the synthe-sis of LEA proteins that prepare the embryo for desiccation.This peak of ABA also initiates seed dormancy.ABA levels fall rapidly in the later stage of seed maturation and are very low in dry seeds. During seed imbibition,de novo ABA biosynthesis in the embryo is a determinant of seed dormancy(Bent-sink and Koornneef,2002;Finkelstein et al.,2002). ABA at this stage also maintains,within a narrow time window,the imbibed embryo in a reversible state between dormancy and germination by regulat-ing the basic Leu Zip transcription factor ABI5 (Lopez-Molina et al.,2001).These important roles of ABA and its dynamics of accumulation in the embryo suggest that ABA biosynthesis is under tight devel-opmental regulation in the embryo.
Transcripts for all the ABA biosynthetic genes are detected in embryos/developing seeds,although more detailed analysis of the expression of individ-ual genes during seed development has not been reported except for AtZEP1.In situ hybridization detected AtZEP expression in the embryo from glob-ular to desiccation stages(Audran et al.,2001),which indicates that the ZEP gene was expressed during embryogenesis before the first peak of ABA accumu-lation in developing seeds.In addition to suggesting a potential role of low-level ABA in embryogenesis,it also raises an interesting question regarding which signal(s)activates ZEP(and other ABA biosynthetic genes)in developing embryos.Although the possi-bility of a unique developmental signal that induces one or more of the ABA biosynthetic genes cannot be ruled out,current experimental evidence implies that soluble sugars,osmotic stress,and ABA itself are likely to be the signals that activate ABA biosynthesis in developing seeds.
Maternal ABA has been shown to be required for the first peak accumulation of ABA in developing seeds,yet it is not clear whether the ABA was directly derived from maternal tissues or rather that maternal ABA serves only as a signal for de novo synthesis of ABA in developing embryos.This question is rele-vant because ABA can positively regulate its own biosynthesis by activating ABA biosynthetic genes (see below).Because ZEP,SDR1,AAO3,and MCSU genes are all induced by sugars to various extents (Cheng et al.,2002),sugar levels may regulate ABA biosynthesis in the embryo.This mechanism is per-haps more important for SDR1because SDR1is not induced by either osmotic stress or ABA.At the onset of seed maturation,osmotic stress may become more important in activating de novo ABA biosynthesis,which is responsible for embryo desiccation tolerance and dormancy.
The activation of individual genes in seeds may eventually be responsible for ABA biosynthesis and accumulation in seeds.For instance,the expression of NtZEP reaches its maximum between one-third and one-half of seed development,which correlates with increased ABA accumulation during this period(Au-dran et al.,1998).Likewise,seeds from plants over-expressing NtZEP had a higher ABA level and showed enhanced dormancy(Frey et al.,1999),sug-gesting that NtZEP may regulate ABA biosynthesis in seeds and during seed germination.Similarly,in tomato,NCED may also regulate ABA levels in the seeds.Overexpression of the LeNCED1gene in-creased ABA levels in imbibed seeds and extended seed dormancy(Thompson et al.,2000b).These ex-perimental data suggest that ABA biosynthesis in developing and imbibing seeds may be regulated at multiple steps.
ABIOTIC STRESS REGULATION OF
ABA BIOSYNTHESIS
Certain environmental signals such as light have been suggested to regulate ABA biosynthesis directly or indirectly.The environmental conditions that most dramatically activate ABA biosynthesis,however, are drought and salt stress.Increased ABA levels under these abiotic stresses result mainly from in-creased de novo biosynthesis.The degradation of ABA appears to be suppressed by stress and acti-vated by ABA and stress relief.
Drought and salt stresses induce ABA biosynthesis largely through transcriptional regulation of ABA biosynthetic genes because blocking transcription by using transcription inhibitors impairs stress-induced ABA biosynthesis.Therefore,transcriptional regula-tion of ABA biosynthetic genes holds the key to understanding how ABA biosynthesis is regulated, although regulation of the specific activities of ABA biosynthesis enzymes also exists.
ZEP was the first gene in the ABA biosynthesis pathway to be cloned,and its expression and regu-lation have been scrutinized in a number of plant species.ZEP genes were expressed ubiquitously in every plant part with a higher basal expression in leaves(Audran et al.,1998;Xiong et al.,2002a).It was thought that ZEP does not limit ABA biosynthesis in photosynthetic tissues because on a molar basis,the amount of9-cis-epoxycarotenoid(precursor down-stream of the ZEP-catalyzed reaction)in photosyn-thetic tissues such as leaves is several times higher than the amount of ABA produced during stress.In tobacco and tomato plants,the transcript levels of ZEP genes in leaves were also not regulated by drought stress but were found to be regulated diur-nally with high transcript levels in the day,which may reflect regulation by the circadian rhythm(Au-
Regulation of Abscisic Acid Biosynthesis
dran et al.,1998;Thompson et al.,2000a)rather than the periods of diurnal water potential changes in plants.Despite the diurnal variation in transcript levels,no change in the level of ZEP protein was found.The potential circadian regulation could be related to the involvement of ZEP products in the light-harvesting complex but not to the role of ZEP proteins in ABA biosynthesis.In contrast to leaves, the amount of epoxycarotenoid precursors is lower in roots,where ZEP,therefore,may limit ABA biosyn-thesis.Consistent with this speculation,ZEP genes in roots were clearly regulated by drought stress.Its transcript levels increased severalfold after drought stress both in tobacco and tomato plants(Audran et al.,1998;Thompson et al.,2000a).
The regulation of ABA biosynthetic genes may vary not only between different plant parts and de-velopmental stages but also between different plant species.Like the ZEP genes in tobacco and tomato, the Arabidopsis ZEP gene also had a basal transcript level under non-stressful conditions.However, drought,salt,and polyethylene glycol clearly in-creased its expression level both in the shoot and in the root(Xiong et al.,2002a),demonstrating that the AtZEP gene is under stress regulation.Variation in the regulation of the ZEP genes observed in different experiments may be partly related to the relative basal transcript levels.A high basal transcript level may mask stress inducibility of the genes. Expression studies with other ABA biosynthetic genes(NCED,AtAAO3,MCSU,and AtSDR1)are less controversial.With the notion that the cleavage step is rate limiting in ABA biosynthesis,the expression of NCED gene(s)has received particular attention. Drought stress treatments were shown to induce NCED expression in maize(Tan et al.,1997),tomato (Burbidge et al.,1999),bean(Phaseolus vulgaris;Qin and Zeevaart,1999),Arabidopsis(Iuchi et al.,2001), cowpea(Vigna unguiculata;Iuchi et al.,2000),and avocado(Persea americana;Chernys and Zeevaart, 2000).Significant increases in NCED transcript levels can be detected within15to30min after leaf detach-ment or dehydration treatment(Qin and Zeevaart, 1999;Thompson et al.,2000a),indicating that the activation of NCED genes can be fairly quick.
In fact,with the exception of AtSDR1,whose ex-pression appears not to be regulated by stress(Cheng et al.,2002;Gonzalez-Guzman et al.,2002),all the other ABA biosynthetic genes are up-regulated by drought and salt stress(Seo et al.,2000b;Iuchi et al., 2001;Xiong et al.,2001b;Xiong et al.,2002a),al-though their protein levels have not been examined in most cases.Because ABA biosynthesis does in-crease dramatically upon stress treatment,it is ex-pected that the protein levels of these genes increase after the transcript levels,as was seen with the NCED gene(Qin and Zeevaart,1999).Nonetheless,a study with AtAAO3suggested that its protein levels did not change as the transcript levels(Seo et al.,2000b).At present,it is not clear whether this is only an exception reflecting the particular experimental con-ditions used.In contrast to the clear regulation of these genes by drought and salt stress,the expression of AtZEP(Xiong et al.,2002a),NCED(Qin and Ze-evaart,1999),and AtMCSU(Xiong et al.,2001b)was not obviously up-regulated by cold.This is consistent with the observation that the magnitude of increase in ABA contents in plants subjected to cold treatment was much less than that in drought-stressed plants (Thomashow,1999).
The unique regulation of the SDR gene is intrigu-ing.AtSDR1(ABA2/GIN1)promoter activity was de-tected mainly in vascular tissues.Thus,its predomi-nant expression tissues appear to be separated from those for other ABA biosynthetic genes that are ex-pressed more ubiquitously(Cheng et al.,2002).The gene is expressed constitutively at a relatively low level and is not induced by drought stress.Rather,its expression is enhanced by sugar.During seed devel-opment,changes in sugar levels in the maturing seeds may have an impact on ABA biosynthesis,as discussed above.Sugar levels also vary diurnally and are influenced by abiotic stress.However,short-term stress treatments under laboratory conditions had no obvious effect on AtSDR1promoter activity(Cheng et al.,2002).
SELF-REGULATION OF ABA
BIOSYNTHETIC GENES
Many biosynthetic pathways are regulated by their end products.ABA has long been thought to nega-tively regulate ABA accumulation by activating its catabolic enzymes(Cutler and Krochko,1999).The activity of a cytochrome P450enzyme,ABA8?-hydroxylase,which catalyzes the first step of ABA degradation,was stimulated by exogenous ABA(e.g. Uknes and Ho,1984).Studies with transgenic tobacco overexpressing NCED gene(or under an inducible promoter)showed that ABA overproduction corre-lated with the overaccumulation of the catabolite, phaseic acid(Qin and Zeevaart,2002).These studies support the notion that ABA might restrict its own accumulation by activating its degradation,at least under non-stressful conditions.On the other hand, whether ABA can stimulate or inhibit its own bio-synthesis was unclear.
Because the NCED gene product has been sug-gested to catalyze the rate-limiting step in the ABA biosynthesis pathway,whether or not this gene is regulated by ABA is very relevant to the question of whether ABA can auto-regulate its own biosynthesis. In tomato plants,it was found that the NCED gene was not induced by exogenous ABA(Thompson et al.,2000a).Similarly,in cowpea,ABA was unable to activate NCED genes(Iuchi et al.,2000).These obser-vations would support the idea that ABA may stim-ulate its own degradation but not its production.
Xiong and Zhu
However,when the expression of ABA biosynthetic genes was examined across wide genetic back-grounds,a different picture emerged.To our sur-prise,we have found that ZEP,AAO3,and MCSU in Arabidopsis are all up-regulated by ABA,in addition to being regulated by stress.Exogenous ABA signif-icantly enhanced the expression of these genes (Xiong et al.,2001a,2001b,2002a).Moreover,these genes appear also to be regulated by endogenous ABA.It was observed that in any of the ABA-deficient mutants los5,aba1,aba2,or aba3,the tran-script levels for all the inducible ABA biosynthetic genes under stress conditions were significantly lower than those in the wild-type plants(Xiong et al., 2002a),although their basal transcript levels were unaffected in these mutants under non-stressful con-ditions(Audran et al.,2001;Xiong et al.,2002a). Thus,ABA deficiency in the Arabidopsis aba mutants may not be simply a consequence of the lesions in the biosynthetic enzymes but also may be because of the significantly reduced expression of other ABA bio-synthetic genes as a result of the primary lesions. Furthermore,it was found that even the NCED gene was induced by ABA in certain genetic back-grounds(e.g.ABA-deficient mutants and certain ecotypes such as Landsberg;Xiong et al.,2002a). Recently,Cheng et al.(2002)also reported that the AtNCED3gene(and AtZEP and AtAAO3)could be induced by ABA in the Landsberg background.In addition,AtNCED3transcript levels under drought and salt stress treatments were significantly reduced in the ABA-deficient mutants los5and los6as com-pared with those in wild-type seedlings,demonstrat-ing that ABA is required for full activation of At-NCED3by osmotic stress(Xiong et al.,2002a).Taken together,these observations strongly suggest a posi-tive feedback regulation of ABA biosynthesis by ABA.This may underscore a stress adaptation mech-anism where an initial induction of ABA biosynthesis rapidly stimulates further biosynthesis of ABA through this positive feedback loop(Fig.1).
To better understand this feedback regulatory cir-cuit,it is important to determine which signaling components may mediate this self-regulation of ABA biosynthetic genes.One such component is SAD1 (supersensitive to ABA and drought1).A recessive mutation in SAD1confers hypersensitivity to exoge-nous ABA during seed germination,vegetative growth,and in gene expression(Xiong et al.,2001a). Interestingly,sad1mutant plants are also impaired in drought-induced ABA biosynthesis.Further charac-terization of sad1found that the mutant was defective in the self-regulatory loop because the sad1mutation impairs ABA regulation of the AAO3and MCSU genes(Xiong et al.,2001a).Both gene products are required for the last step of ABA biosynthesis,i.e.the conversion of ABA aldehyde to ABA(Fig.1).Signif-icantly,a feeding assay showed that the conversion of ABA-aldehyde to ABA was impaired in the sad1mutant(Xiong et al.,2001a).This defect may well be responsible for the impaired drought-induced ABA biosynthesis in the mutant.It is likely that SAD1,an Sm-like small ribonucleoprotein that is predicted to be involved in mRNA splicing,export,and degrada-tion,may regulate the turnover rates for the tran-scripts of an early ABA signaling component(s).The components(s)may in turn regulate the feedback circuit.One such component is likely to be an ABI1-like protein phosphatase2C because stress induction of this gene was impaired in sad1(Xiong et al.,2001a). Although a mutation in the mRNA cap-binding pro-tein also reduced the expression of another PP2C gene,this ABA-hypersensitive mutant,abh1,was not ABA deficient(Hugouvieux et al.,2001).Thus,at least some of the PP2Cs,including ABI1itself,may be involved in this self-regulation circuit as discussed below.
Arabidopsis ABI1and ABI2are homologous2C-type protein phosphatases that may negatively reg-ulate ABA signaling.The abi1-1and abi2-1mutations confer ABA insensitivity in seed germination,vege-tative growth,and the expression of certain ABA-regulated genes(Finkelstein et al.,2002).In abi1,the self-regulation loop of ABA biosynthesis is partially impaired because in this mutant,ABA fails to acti-vate the expression of the NCED gene(in Landsberg background),and this mutation significantly reduced the transcript levels of ZEP and AAO3under ABA treatment.Strikingly,all these genes were unaffected in abi2(Xiong et al.,2002a).Despite their sequence homology and overlapping functions,ABI1and ABI2 are often found to have different roles in ABA sig-naling.For example,it was shown that ABA-induced reactive oxygen species(ROS)production was im-paired in abi1but not in abi2.In contrast,hydrogen peroxide activates plasma membrane Ca2?channels and induces stomatal closure in abi1but not in abi2 (Murata et al.,2001).Because ROS is involved in ABA signaling and probably also in regulating ABA bio-synthesis(Zhao et al.,2001),it is possible that this self-regulation of ABA biosynthetic genes may be partly mediated by ROS through a protein phosphor-ylation cascade(Fig.1).In any case,the involvement of ABI1in the feedback loop further suggests that signaling for ABA biosynthesis is ABA dependent and that there is cross talk between the signaling pathway for ABA biosynthesis and the pathway for ABA responsiveness.Future studies are needed to identify the interplay between these two pathways and the significance of self-regulation of ABA biosyn-thesis in overall ABA accumulation under abiotic stress conditions.
DIFFERENTIAL REGULATION OF ABA BIOSYNTHETIC GENES AND THE RATE-LIMITING STEP IN THE BIOSYNTHESIS PATHWAY
Genes involved in ABA biosynthesis exist either as a single copy or a gene family,and the family mem-
Regulation of Abscisic Acid Biosynthesis
bers may be subjected to differential regulation as discussed above.For Arabidopsis,whose genome has been completely sequenced,the copy numbers of ABA biosynthetic genes are known(Table I).It seems that ZEP,MCSU,and SDR are single-copy genes, whereas NCED and AAO belong to gene families. Current data suggest that for those belonging to gene families,each member is regulated differently by stresses.In addition,they may be expressed in a tissue-and developmental stage-specific manner.For example,AtAAO3was expressed in leaves but not in the root,and AAO4was mainly expressed in siliques (Seo et al.,2000a),whereas AtAAO3may be less expressed in seeds.Thus,aao3mutant seeds were not obviously changed in dormancy(Seo et al.,2000b), which is clearly different from other known ABA-deficient mutants whose seeds are much less dor-mant.This may explain why aao3mutants were not isolated in germination screens.It is also likely that family members may be regulated differentially by different stresses or may have different threshold of stress induction.
Because the ABA biosynthesis pathway involves multiple gene products,there could be a rate-limiting step in the pathway.Finding this limiting step is important for genetic manipulation of the pathway. With the complex regulation mechanisms for indi-vidual ABA biosynthetic genes that may vary among plant species,together with the mobile nature of ABA or its immediate precursors,it would be diffi-cult to draw a consensus regarding the regulatory patterns of these genes or the rate-limiting step for ABA biosynthesis in the whole plant.Nonetheless,it was recognized generally that the step catalyzed by NCED,i.e.the oxidative cleavage of neoxanthin,is rate limiting(Tan et al.,1997;Qin and Zeevaart,1999; Taylor et al.,2000;Thompson et al.,2000b).This may be valid in leaves,where most studies on ABA bio-synthesis are concerned.Consistent with this predic-tion,constitutive or inducible overexpression of the NCED genes resulted in increased ABA biosynthesis and reduced transpiration water loss(Iuchi et al., 2001;Qin and Zeevaart,2002;Thompson et al., 2000b).However,if the rate-limiting step is solely at the NCED,then one would expect that overexpres-sion of other ABA biosynthetic genes would not sig-nificantly increase ABA biosynthesis,at least not in the leaves.Nonetheless,experimental evidence indi-cated that even overexpressing the AtZEP gene, whose product catalyzes the least rate-limiting step considered in the pathway,could result in an in-creased stress gene induction in Arabidopsis seed-lings(Xiong et al.,2002a).Furthermore,overexpres-sion of NtZEP led to increased seed dormancy and delayed seed germination in tobacco(Frey et al., 1999),presumably as a result of increased ABA bio-synthesis in imbibed embryos.These observations would suggest that ZEP might limit ABA biosynthe-sis also,particularly in non-photosynthetic tissues such as seeds and roots.It would be interesting to see whether up-regulating the expression level of other ABA biosynthetic genes can cause enhanced ABA biosynthesis.
The possibility that overexpressing any of the ABA biosynthetic genes probably have an impact on ABA biosynthesis may in fact have to do with the self-regulation mechanism discussed in the previous sec-tion.This is because a limited initial increase in ABA biosynthesis from overexpressing a single ABA bio-synthetic gene may result in a coordinately increased induction of other ABA biosynthetic genes through the self-regulatory loop(Fig.1).This possibility can be confirmed by examining the expression of other ABA biosynthetic genes in the overexpressors.In
Table I.ABA biosynthetic genes and their regulation in Arabidopsis
Not all the genes listed are involved in ABA biosynthesis;those known to be involved are indicated in bold.Gene symbols are given in Figure1.
Genes Arabidopsis Genome Initative
Identification
Mutants Expression Pattern Stress Inducibility
ZEP At5g67030aba1,los6,npq2Ubiquitous Inducible in roots,
also in leaves
SDR1At1g52340aba2,gin1,sis4,
isi4,sre1,san3Ubiquitous?Vascular
tissues
Not induced by
stress but by sugar
NCED1/CCD1At3g63520Not inducible? NCED2At4g18350Not inducible NCED3At3g14440nced3Ubiquitous?Strongly inducible NCED4At4g19170Not inducible NCED5At1g78390Not inducible NCED6At3g24220Not inducible NCED9At1g30100Weakly inducible AAO1At5g20960Roots and seeds
AAO2At3g43600Roots
AAO3At2g27150aao3Leaves but not siliques/
seeds Inducible in leaves but not in roots
AAO4At1g04580Mainly in siliques
MCSU At1g16540aba3,los5,frs1Ubiquitous Inducible Xiong and Zhu
addition,the fact that NCED s are either not up-regulated or weakly up-regulated by ABA may cause the NCED step to become rate limiting late in ABA biosynthesis in leaves.As a consequence,regulating NCED genes may have a more significant impact on overall ABA biosynthesis.
UNRAVELING THE REGULATORY CIRCUIT FOR ABA BIOSYNTHESIS
Obviously,the regulation of ABA biosynthesis is of great importance in controlling ABA level and fine-tuning plant stress responses and developmental programs.Because stress-inducible ABA biosynthetic genes contain both the DRE-and ABRE-like cis-elements in their promoters(Xiong et al.,2001b;Bray, 2002),it is possible,therefore,that these genes may be similarly regulated as the DRE/CRT class of stress-responsive genes(Xiong et al.,2002b).Circum-stantial evidence has suggested that the signal trans-duction pathways for stress-induced ABA biosynthe-sis may involve redox signals,Ca2?signaling,and protein phosphorylation and dephosphorylation events.Yet,biochemical or genetic studies of each of these signaling processes in relation to ABA biosyn-thesis have been lacking.In the near future,molecu-lar studies should identify the cis-elements in ABA biosynthetic genes and the respective transcription factors that are responsible for the activation of these genes.Extensive genetic analysis will also be needed to dissect such a complex signal transduction process.
Exhaustive screens of seed germination,either in the presence of GA synthesis inhibitors or high con-centrations of salt,have identified lesions in ABA biosynthesis(and in ABA responsiveness).These mutants also were repeatedly recovered in screens for sensitivity to sugars(Finkelstein et al.,2002;Rol-land et al.,2002).In fact,some loci were identified in more than six independent screening schemes!None-theless,no component that directly regulates ABA biosynthesis has emerged in these screens.Because sugar,ethylene,and ABA biosynthesis have a com-plicated interplay in seed germination and in some other physiological processes(Ghassemian et al., 2000;Hansen and Grossmann,2000;Gonzalez-Guzman et al.,2002),there is a possibility that some of the existing signaling components in other hor-mone response pathways may be involved in ABA biosynthesis regulation but have escaped our atten-tion.One such example may be era3(enhanced re-sponse to ABA3)/ein2(ethylene insensitive2).era3/ein2 plants have a basal ABA level twice of that in the wild type,which is probably due to increased bio-synthesis because the ZEP transcript level was also higher in ein2(Ghassemian et al.,2000).Yet,the effect of the ein2mutation on ABA biosynthesis could be a consequence of the complex interaction between dif-ferent hormones rather than a specific regulation of ABA biosynthesis.Aside from possible genetic re-dundancy of the regulatory pathways,the difficulty in identifying regulatory loci in seed-based screens may also have to do with the possibility that these screens are not suitable for the identification of the components.To uncover the regulatory pathways for ABA biosynthesis,one may need additional approaches.
Given the roles of ABA in stress responses at the vegetative stage,screens based on stress responses in vegetative tissues would likely identify new loci that are important in regulating ABA biosynthesis or sig-naling.Due to the difficulty in manipulating drought stress in a quantitative and reproducible way,screens for altered drought tolerance have not been widely used.Alternative screens such as those for guard cell regulation(e.g.Mustilli et al.,2002)and gas exchange (e.g.sensitivity to CO2)may discover novel loci that regulate ABA accumulation or signaling.Because gene expression is more sensitive to stress regulation than are some of the visible phenotypes,molecular genetic approaches such as the one used in the screen for stress signal transduction mutants(Ishitani et al., 1997)may be more productive in identifying signal transduction components.In this approach,the pro-moters of stress-inducible ABA biosynthetic genes(in particular,AtNCED3,AtMCSU,and AtAAO3)can be transcriptionally fused to a reporter gene;then,mu-tants with altered reporter gene expressions in re-sponse to abiotic stress can be isolated.These screens may uncover novel regulatory mechanisms in ABA biosynthesis.With the availability of the complete genomic information of Arabidopsis and large bodies of expression data,reverse genetics approaches should also facilitate the identification of new regu-latory components in the signaling pathway leading to ABA biosynthesis.A complete understanding of the regulation of ABA biosynthesis will require a combination of genetics,genomics,molecular biol-ogy,and biochemical approaches.
Received April12,2003;returned for revision May12,2003;accepted May 22,2003.
LITERATURE CITED
Audran C,Borel C,Frey A,Sotta B,Meyer C,Simonneau T,Marion-Poll A(1998)Expression studies of the zeaxanthin epoxidase gene in Nicotiana plumbaginifolia.Plant Physiol118:1021–1028
Audran C,Liotenberg S,Gonneau M,North H,Frey A,Tap-Waksman K, Vartanian N,Marion-Poll A(2001)Localisation and expression of zeax-anthin epoxidase mRNA in Arabidopsis in response to drought stress and during seed development.Aust J Plant Physiol28:1161–1173 Bentsink L,Koornneef M(2002)Seed dormancy and germination.In CR Somerville,EM Meyerowitz,The Arabidopsis Book.American Society of Plant Biologists,Rockville,MD,pp1–17.https://www.wendangku.net/doc/bb4899823.html,/publica-tions/Arabidopsis
Bittner F,Oreb M,Mendel RR(2001)ABA3is a molybdenum cofactor sulfurase required for activation of aldehyde oxidase and xanthine de-hydrogenase in Arabidopsis thaliana.J Biol Chem276:40381–40384
Bray EA(2002)Abscisic acid regulation of gene expression during water-deficit stress in the era of the Arabidopsis genome.Plant Cell Environ25: 153–161
Regulation of Abscisic Acid Biosynthesis
Burbidge A,Grieve TM,Jackson A,Thompson A,McCarty DR,Taylor IB (1999)Characterization of the ABA-deficient tomato mutant notabilis and its relationship with maize vp14.Plant J17:427–431
Cheng WH,Endo A,Zhou L,Penney J,Chen HC,Arroyo A,Leon P, Nambara E,Asami T,Seo M et al.(2002)A unique short-chain dehy-drogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions.Plant Cell14:2723–2743
Chernys JT,Zeevaart JA(2000)Characterization of the9-cis-epoxycarotenoid dioxygenase gene family and the regulation of abscisic acid biosynthesis in avocado.Plant Physiol124:343–353
Cutler A,Krochko J(1999)Formation and breakdown of ABA.Trends Plant Sci4:472–478
Finkelstein RR,Gampala SS,Rock CD(2002)Abscisic acid signaling in seeds and seedlings.Plant Cell14:S15–S45
Finkelstein RR,Rock CD(2002)Abscisic acid biosynthesis and response.In CR Somerville,EM Meyerowitz,eds,The Arabidopsis Book.American Society of Plant Biologists,Rockville,MD,pp1–52.http://www.aspb.
org/publications/Arabidopsis
Frey A,Audran C,Marin E,Sotta B,Marion-Poll A(1999)Engineering seed dormancy by the modification of zeaxanthin epoxidase gene expression.
Plant Mol Biol39:1267–1274
Ghassemian M,Nambara E,Cutler S,Kawaide H,Kamiya Y,McCourt P (2000)Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis.Plant Cell12:1117–1126
Gonzalez-Guzman M,Apostolova N,Belles JM,Barrero JM,Piqueras P, Ponce MR,Micol JL,Serrano R,Rodriguez PL(2002)The short-chain alcohol dehydrogenase ABA2catalyzes the conversion of xanthoxin to abscisic aldehyde.Plant Cell14:1833–1846
Hansen H,Grossmann K(2000)Auxin-induced ethylene triggers abscisic acid biosynthesis and growth inhibition.Plant Physiol124:1437–1448 Hasegawa PM,Bressan RA,Zhu JK,Bohnert HJ(2000)Plant cellular and molecular responses to high salinity.Annu Rev Plant Mol Plant Physiol 51:463–499
Hugouvieux V,Kwak JM,Schroeder JI(2001)An mRNA cap binding protein,ABH1,modulates early abscisic acid signal transduction.Cell 106:477–487
Ishitani M,Xiong L,Stevenson B,Zhu J-K(1997)Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis:interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways.Plant Cell9:1935–1949
Iuchi S,Kobayashi M,Taji T,Naramoto M,Seki M,Kato T,Tabata S, Kakubari Y,Yamaguchi-Shinozaki K,Shinozaki K(2001)Regulation of drought tolerance by gene manipulation of9-cis-epoxycarotenoid dioxy-genase,a key enzyme in abscisic acid biosynthesis in Arabidopsis.Plant J27:325–333
Iuchi S,Kobayashi M,Yamaguchi-Shinozaki K,Shinozaki K(2000)A stress-inducible gene for9-cis-epoxycarotenoid dioxygenase involved in abscisic acid biosynthesis under water stress in drought-tolerant cowpea.
Plant Physiol123:553–562
Lopez-Molina L,Mongrand S,Chua NH(2001)A postgermination devel-opmental arrest checkpoint is mediated by abscisic acid and requires the ABI5transcription factor in Arabidopsis.Proc Natl Acad Sci USA98: 4782–4787
Marin E,Nussaume L,Quesada A,Gonneau M,Sotta B,Hugueney P,Frey A,Marion-Poll A(1996)Molecular identification of zeaxanthin epoxi-dase of Nicotiana plumbaginifolia,a gene involved in abscisic acid biosyn-thesis and corresponding to the ABA locus of Arabidopsis thaliana.EMBO J15:2331–2342
Murata Y,Pei ZM,Mori IC,Schroeder JI(2001)Abscisic acid activation of plasma membrane Ca2?channels in guard cells require cytosolic NAD(P)H and is differentially disrupted upstream and downstream of reactive oxygen species production in abi1-1and abi2-1protein phospha-tase2C mutants.Plant Cell13:2513–2523
Mustilli AC,Merlot S,Vavasseur A,Fenzi F,Giraudat J(2002)Arabidopsis OST1protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production.
Plant Cell14:3089–3099Phillips J,Artsaenko O,Fiedler U,Horstmann C,Mock H-P,Muntz K, Conrad U(1997)Seed-specific immunomodulation of abscisic acid activ-ity induces a developmental switch.EMBO J16:4489–4496
Qin X,Zeevaart JAD(1999)The9-cis-epoxycarotenoid cleavage reaction is the key regulatory step of abscisic acid biosynthesis in water-stressed bean.Proc Natl Acad Sci USA96:15354–15361
Qin X,Zeevaart JAD(2002)Overexpression of a9-cis-epoxycarotenoid dioxygenase gene in Nicotiana plumbaginifolia increases abscisic acid and phaseic acid levels and enhances drought tolerance.Plant Physiol128: 544–551
Rolland F,Moore B,Sheen J(2002)Sugar sensing and signaling in plants.
Plant Cell14:S185–205
Rook F,Corke F,Card R,Munz G,Smith C,Bevan MW(2001)Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signaling.Plant J26: 421–433
Schwartz SH,Qin X,Zeevaart JAD(2003)Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants,genes,and enzymes.
Plant Physiol131:1591–1601
Schwartz SH,Tan BC,Gage DA,Zeevaart JAD,McCarty DR(1997)Spe-cific oxidative cleavage of carotenoid by VP14of maize.Science276: 1872–1874
Seo M,Koiwa H,Akaba S,Komano T,Oritani T,Kamiya Y,Koshiba T (2000a)Abscisic acid aldehyde oxidase of Arabidopsis thaliana.Plant J23: 481–488
Seo M,Koshiba T(2002)Complex regulation of ABA biosynthesis in plants.
Trends Plant Sci7:41–48
Seo M,Peeters AJM,Koiwai H,Oritani T,Marion-Poll A,Zeevaart JAD, Koorneef M,Kamiya Y,Koshiba T(2000b)The Arabidopsis aldehyde oxidase3(AAO3)gene product catalyzes the final step in abscisic acid biosynthesis in leaves.Proc Natl Acad Sci USA97:12908–12913
Sharp RE(2002)Interaction with ethylene:changing views on the role of abscisic acid in root and shoot growth responses to water stress.Plant Cell Environ25:211–222
Tan BC,Schwartz SH,Zeevaart JAD,McCarty DR(1997)Genetic control of abscisic acid biosynthesis in maize.Proc Natl Acad Sci USA94: 12235–12240
Taylor IB,Burbidage A,Thompson AJ(2000)Control of abscisic acid synthesis.J Exp Bot51:1563–1574
Thomashow MF(1999)Plant cold acclimation:freezing tolerance genes and regulatory mechanisms.Annu Rev Plant Physiol Plant Mol Biol50: 571–599
Thompson AJ,Jackson AC,Parker RA,Morpeth DR,Burbidge A,Taylor IB(2000a)Abscisic acid biosynthesis in tomato:regulation of zeaxanthin epoxidase and9-cis-epoxycarotenoid dioxygenase mRNAs by light/dark cycles,water stress and abscisic acid.Plant Mol Biol42:833–845 Thompson AJ,Jackson AC,Symonds RC,Mulholland BJ,Dadswell AR, Blake PS,Burbidge A,Taylor IB(2000b)Ectopic expression of a tomato 9-cis-epoxycarotenoid dioxygenase gene causes over-production of ab-scisic acid.Plant J23:363–374
Uknes SJ,Ho THD(1984)Mode of action of abscisic acid in barley aleurone layers.Abscisic acid induces its own conversion to phaseic acid.Plant Physiol75:1126–1132
Xiong L,Gong Z,Rock C,Subramanian S,Guo Y,Xu W,Galbraith D,Zhu JK(2001a)Modulation of abscisic acid signal transduction and biosyn-thesis by an Sm-like protein in Arabidopsis.Dev Cell1:771–781
Xiong L,Ishitani M,Lee H,Zhu JK(2001b)The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold and osmotic stress-responsive gene expression.Plant Cell13:2063–2083 Xiong L,Lee H,Ishitani M,Zhu JK(2002a)Regulation of osmotic stress-responsive gene expression by the LOS6/ABA1locus in Arabidopsis.
J Biol Chem277:8588–8596
Xiong L,Shumaker KS,Zhu JK(2002b)Cell signaling during cold,drought and salt stresses.Plant Cell14:S165–S183
Zhao Z,Chen G,Zhang C(2001)Interaction between reactive oxygen species and nitric oxide in drought-induced abscisic acid synthesis in root tips of wheat seedlings.Aust J Plant Physiol28:1055–1061
Xiong and Zhu
